Environmental DNA Reveals Surprising Shark Diversity

Dr. Bakker samples water with sharks (Credit: screenshot, University of Salford, https://www.youtube.com/watch?v=ucj8Ftgvwdg)
Recent work from marine ecologists reveals that the power of environmental DNA (eDNA) extends beyond shorelines and deep into the Earth’s oceans, revealing that vulnerable, rare shark species still live in regions where they were believed to be extinct. Their work also illustrates both the potentials and pitfalls of working with eDNA in the water.
Seeking out elusive marine species
Some marine species—sharks being a prime example—are big, elusive, highly mobile, and rare. They avoid humans with good reason; we often compete for the same resources, and we’ve driven many species toward the brink of extinction. Furthermore, sharks are tough to trap, and potentially aggressive when taken.
These marine megafaunas are in serious decline around the world. This dangerous dip in marine biodiversity is cloaked in guesswork for scientists, because detecting species like these is challenging, at best, even for experts. This prompts the question: how much of the decline of a species like sharks is real, and how much is down to our inability to detect and track them effectively?
The team in this international study, composed of scientists in France, New Caledonia, and the UK, found they were able to detect residual populations of multiple species of sharks using eDNA, which is shed from blood and other excretions as well as skin. This success was especially notable because the species had gone undetected in the areas in question for years.
“I was inspired by reading Thomsen et al. from 2012, plain and simple,” comments conservation genetics professor and study co-leader Dr. Stefano Mariani of the University of Salford. “I thought: ‘Wow, this is like ocean forensics.’”
Along with colleague Judith Bakker, PhD, also at the University of Salford, Dr. Mariani and the team set out to collect samples of their own and validate their theory: that eDNA could help them sharpen conservation goals and augment our understanding of shark and ray biodiversity.

Dr. Mariani takes a sample. (Credit: screenshot, University of Salford, https://www.youtube.com/watch?v=ucj8Ftgvwdg)
Metabarcoding detectives
Typically, researchers study DNA after extracting it from a blood sample or a piece of tissue. However, as anyone who’s seen a (frequently misleading!) episode of CSI knows, humans shed DNA all the time, and other animals are no different. In the ocean, eating, fighting, urinating, mating and dying, cause animals to release DNA into the environment. This trace or eDNA gives scientists clues as to which species deposited it, and how.
The team hoped that they could sample ocean water and sequence DNA fragments it carried to determine where and how sharks were distributed in a region, quickly and easily. They set out to measure the eDNA technique’s efficiency for shark monitoring by contrasting its performance with proven methodologies. In this case, the comparative techniques were visual censuses from scuba-diving and recordings from baited underwater cameras (BRUVs).
The team used a process called metabarcoding to analyze the water samples. This non-invasive approach is based on the use of universal primer pairs, group-specific polymerase chain reaction (PCR) DNA sequencing primers that bind to a commonly found sequence.
They were compelling: in a few weeks of collecting the team took 22 water samples, and detected 13 shark species—more than the nine species that had been seen over the course of more than 3,000 dives in the same regions for decades. Similarly, it took almost 400 BRUVs observations over two years to match the number of species detected in the eDNA analysis.
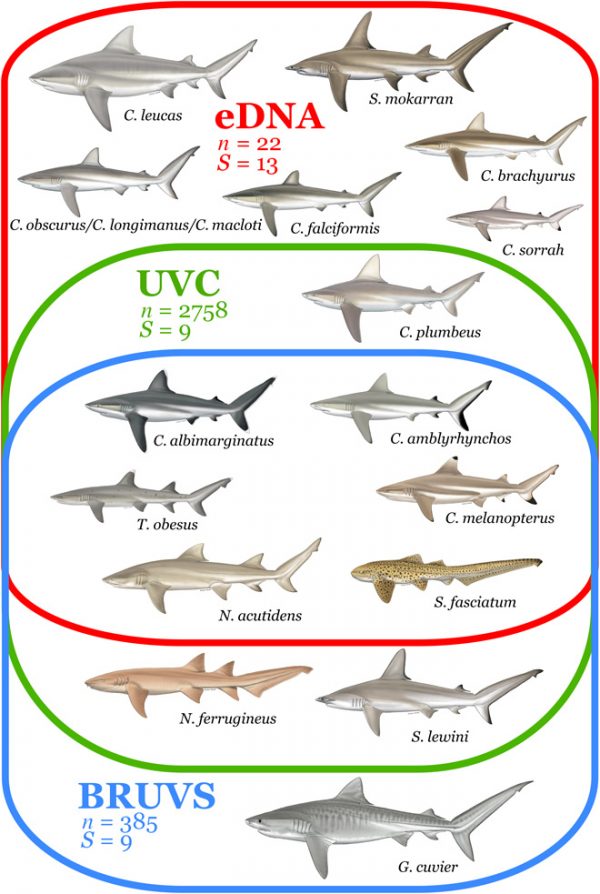
Detection of shark species with different sampling methods. (Credit: Boussarie et al., http://advances.sciencemag.org/content/4/5/eaap9661.full.)
Moreover, almost half of the species the team detected using eDNA were entirely undetectable using traditional methods. Overall, eDNA detected some sharks in approximately 90 percent of the samples. This was a stark contrast to the 50 percent rate for BRUVs, and 15 percent found via scuba diving.
“It is fairly intuitive to imagine that if you go in the water and swim around with gurgling sounds, animals will be suspicious,” remarks Dr. Mariani. “It’s a bit less intrusive to drop a weird clangy thing with some food. But surely sifting minute biological residues from water samples is the least invasive method, no?”
Using eDNA in water is a tricky proposition, whether scientists are working to determine how far benthic creatures are roaming in a river, or which waters sharks are swimming in. Understanding the limitations of the technique is essential to interpreting results.
“It is a matter of scale: the ocean is massive compared to a river, it is also multidimensional, and currents are more complicated, so if anything, doing this at sea is more challenging than in a river,” details Dr. Mariani. “However, if you talk about large beasts that could travel hundreds of miles in a few days, and you are not interested in small, patchy microhabitats, then you can have a broad-sweep idea of what travels in those waters. Obviously, we are far from being able to assign specific, localized, preferred areas of dwelling, because nobody has yet included models of oceanographic circulation in eDNA spatial patterns. And in any case, there is a need for a much heavier sampling/screening effort, both spatially and temporally.”
And while the team was not able to directly confirm that the DNA of the additional four species that had not been seen or recorded otherwise was recent enough to be a sign of live specimens, the nature of eDNA makes that conclusion fairly obvious.

The sampler collects water as a shark swims nearby. (Credit: screenshot, University of Salford, https://www.youtube.com/watch?v=ucj8Ftgvwdg)
“We cannot really have the direct proof of that, but we know that DNA in the water column degrades beyond the limits of detectability within a couple of days, so it would be very awkwardly unlikely that there were recently-dead carcasses, or fresh pieces, of each one of those species in proximity of the sampling,” states Dr. Mariani. “Furthermore, even if that was the case, that would probably mean that there are a few of those animals around anyway.”
Conservation with eDNA
The results were consistent for both highly impacted coastal areas and remote, pristine wilderness areas. In other words, eDNA analysis is powerful enough to detect elusive megafauna species even in depleted areas previously assumed to be devoid of sharks. It also provided a hopeful sense of biodiversity, since a single dive revealed three species of sharks a most, whereas the team detected as many as seven species in a single sample.
The results also suggest that there may be small numbers of certain species we previously thought were gone, that remain in various places—species that are in need of protection. The eDNA approach can assist conservation efforts by assessing the true range of threatened species and presenting a truer picture of biodiversity patterns across large habitats, over time, over a range of environmental gradients. Almost half of all species of sharks, rays, and chimeras on the IUCN Red List have an undetermined protection status because there is insufficient data to determine their numbers and range; this study shows that eDNA may solve this problem.
Of course, eDNA science is still changing and growing. Without reference samples, there’s no way to match eDNA found in samples to what’s in the database. However, scientists all over the world are adding new references to the database every day.
Furthermore, researchers are working to lengthen existing DNA probes, to better distinguish between similar species; Dr. Mariani’s team is already working on the problem.

Processing the samples for DNA. (Credit: screenshot, University of Salford, https://www.youtube.com/watch?v=ucj8Ftgvwdg)
“The scientific community is constantly updating the database so that it being taken care of gradually,” explains Dr. Mariani. “Our priority now is to obtain many more samples of water, from many more reef locations, in order to have a much more robust understanding of this approach and, in doing so, make our results more relevant to active conservation planning.”
Dr. Mariani also has advice for anyone learning about the team’s research:
“I would say, to paraphrase a famous shark scientist: ‘Be skeptical… but open-minded!’”
See more about the work in this video.
Top image: Dr. Bakker samples water with sharks (Credit: screenshot, University of Salford, https://www.youtube.com/watch?v=ucj8Ftgvwd)




0 comments